Telangana TSBIE TS Inter 2nd Year Botany Study Material 10th Lesson Molecular Basis of Inheritance Textbook Questions and Answers.
TS Inter 2nd Year Botany Study Material 10th Lesson Molecular Basis of Inheritance
Very Short Answer Type Questions
Question 1.
What is the function of histones in DNA packaging?
Answer:
- Histones are positively charged basic proteins. They are organized to form a unit of eight molecules called histone octamer.
- The negatively charged DNA is wrapped around the positively charged histone octamer to form a structure called nucleOsome.
Question 2.
Distinguish between heterochromatin and euchromatin. Which of the two is transcriptionally active? [Mar. 2020]
Answer:
1. Euchromation :
Regions of chromatin that are loosely packed and stained lightly. It is transcriptionally active.
2. Heterochromation :
Regions of chromatin that are densely packed and stained dark. It is transcriptionally inactive.
Question 3.
Who proved that DNA is genetic material? What is the organism they worked on? [May 2017]
Answer:
- Alfred Hershey and Martha Chase proved that DNA is genetic material.
- They worked with viruses that infect bacteria, bacteriophages.
Question 4.
What is the function of DNA polymerase?
Answer:
- DNA polymerase uses a DNA template to catalyze polymerization of deoxynucleotides.
- It is highly efficient and catalyses polymerization in only one direction (5′ → 3′) with high degree of accuracy.
Question 5.
What are the components of a nucleotide? [Mar. ’18, ’17; May ’14]
Answer:
- A pentose sugar, a phosphate group and a nitrogen base are the 3 components of a nucleotide.
- The pentose sugar is ribose in nucleotides of RNA and it is deoxyribose in nucleotides of DNA.
![]()
Question 6.
Write the structure of chromatin.
Answer:
- Nucleosomes are formed due to wrapping of negatively charged DNA around the positively charged Histone octamers. These repeating units in the nucleus form chromatin.
- The nucleosomes in chromatin are seen as ‘beads – on – string’.
Question 7.
What is the cause of discontinuous synthesis of DNA on one of its parental strands? What happens to these short stretches of synthesised DNA?
Answer:
- The DNA – dependent DNA polymerase can catalyze polymerization in only one direction, 5′ → 3′ and hence DNA synthesis is discontinuous on lagging of strand (with 5′ → 3′ polarity) of parental DNA.
- The discontinuously synthesized Okazaki fragments on lagging strand are later joined by DNA ligase to form a continuous strand.
Question 8.
Given below is the sequence of coding strand of DNA in a transcription unit 3′ – AATGCAGCTATTAGG – 5′
Write the sequence of
a) its complementary strand
b) the mRNA
Answer:
a) Its complementary strand
5′ – TTACGTCGATAATCC – 3′
b) The mRNA
5′- UUACGUCGAUAAUCC – 3′
Question 9.
In a nucleus, the number of ribonucleoside triphosphates is 10 times the number of deoxy ribonucleoside triphosphates, but only deoxyribonucleotides are addded during the DNA replication. Suggest a mechanism.
Answer:
- DNA is a polymer made of deoxyribonucleotides.
- Absence of 2 – OH group is nuclosides confers stability to DNA molecules.
Question 10.
Name any three viruses which have RNA as the genetic material.
Answer:
- Tobacco Mosaic virus
- Polio virus
- HIV
- QB Bacteriophage
Question 11.
Write the sequence of nucleotides after single base insertion and deletion in the given gene:
Gene: THE CAT ATE THE FAT RAT
Answer:
1. If single base T is inserted between ‘THE’ and ‘CAT’, then
Gene : THE TCA TAT ETH EFA TRA T
2. If single base C is deleted from ‘CAT’, then
Gene: THE ATA TET HEF ATR AT
Question 12.
Why was DNA chosen over RNA as genetic material in the majority of the organisms?
Answer:
- RNA consists of 2 – OH’ group at every nucleotide which is reactive group and makes RNA liable and eassily degradable. It is single stranded, catalyst, reactive and hence unstable.
- DNA lacks 2 – OH’ group, double stranded, consists of thymine and resists changes by evolving a process of repair. Hence it is a stable genetic material in majority of the organisms.
Question 13.
What are the components of a transcription unit? [March 2019]
Answer:
The three major components of a transcription unit are (1) A Promoter (2) The structural gene (3) A terminator.
![]()
Question 14.
What is the difference between exons and introns?
Answer:
1. Exons :
The coding (expressed sequences in split genes of eukaryotes and will appear in processed (matured) RNA.
2. Introns :
The non coding sequences in split genes of eukaryotes that interrupt introns and they do not appear in processed RNA.
Question 15.
What is meant by capping and tailing? [May 2017]
Answer:
1. Capping :
It is a process in which, an unusual nucleotide (methyl guanosine triphosphate) is added to the 5′ end of hn RNA.
2. Tailing :
In this process, adenylate residues (200 – 300) are added at 3′ – end in a template independent manner.
Question 16.
What is meant by point mutation? Give an example. [May 2014]
Answer:
- The mutation that occurs in a single base pair of a gene is called point (gene) mutation.
- A point mutation in the gene for Beta globin chain (in human haemoglobin) results a disease, sickle cell anaemia.
Question 17.
Define peptide bond. Why are proteins referred to as polypeptide chains?
Answer:
- The bond between two adjacent amino acids in a protein is known as peptide bond.
- Proteins are the macromolecules and polymers. Amino acids, are joined by many peptide bonds to form proteins and hence are referred as polypeptide chains.
Question 18.
What is meant by charging of tRNA?
Answer:
- Charging of tRNA (amino acylation of tRNA): Amino acids are activated in the presence of ATP and linked to their cognate tRNAs.
- This favours the formation of peptide bond by providing energy.
Question 19.
What is a regulator and a promoter?
Answer:
- Regulator : This gene directs the activity of the operator gene by producing an inhibitor protein called repressor.
- Promoter is a region of DNA where RNA polymerase binds and initiates transcription.
![]()
Question 20.
During DNA replication, why is it that the entire molecule does not open in one go? Explain replication fork.
Answer:
- For long DNA molecules, the entire molecule does not open in one go due to very high energy requirement.
- The replication occur within a small opening of the DNA helix referred to as replication fork. This is the Y-shaped structure formed when the double-stranded DNA is unwounded upto a point during its replication.
Question 21.
What is the function of the codon-AUG? [Mar. ’20, ’14]
Answer:
- AUG acts as the initiation codon (start codon) during formation of mRNA.
- It also codes for an aminoacid, Methionine.
Question 22.
Define stop codon. Write the codons. [March 2019]
Answer:
- The codons that do not code for any amino acid and responsible for termination of protein synthesis / translation process are called as stop codons.
- There are 3 stop codons ie., UAA, UAG and UGA.
Question 23.
What is the difference between the template strand and a coding strand in a DNA molecule? [May 2014]
Answer:
| Template strand | Coding strand |
| 1) The strand with 3′ → 5′ polarity is a replicating DNA. | 1) It is a strand with 5′ → 3′ polarity in a replicating DNA. |
| 2) This consists of structural gene flanked by promoter and terminator sequences respected at 3′ and 5′ ends. | 2) It has terminator towards 3′ end. |
![]()
Question 24
Write any two chemical differences between DNA and RNA. [March 2017]
Answer:
| DNA | RNA |
| 1) DNA has deoxyribose sugar. | 1) RNA has Ribose sugar. |
| 2) It has thymine and cytosine as pyrimidine bases. | 2) It has uracil and cytosine as pyramidine base. |
Question 25.
In a typical DNA molecule, the proportion of Thymine is 30% of the N bases. Find out the percentages of other N bases.
Answer:
- According to Erwin Chargaff in double stranded DNA the ratio between A and T and between G and C are constant and each equals one.
- Adenine – 30%, Guanine – 20%, Cytocine – 20%.
Question 26.
The proportion of nucleotides in a given nucleic acid are: Adenine 18%, Guanine 30%, Cytosine 42%, and Uracil 10%. Name the nucleic acid and mention the number of strands in it. [March 2018]
Answer:
- As there is Uracil (pyramidine) is the given problem the nucleic acid is RNA.
- The proportion of A ≠ U and G ≠ C, so it is single stranded RNA.
Question 27.
If the base sequence of a codon in mRNA is 5′ AUG-3′. What is the sequence of tRNA pairing with it?
Answer:
3′ – UAC – 5′
Short Answer Type Questions
Question 1.
Define transformation in Griffith’s experiment. Discuss how it helps in the identification of DNA as genetic material.
Answer:
Transformation is defined as the uptake of a naked DNA molecule of the fragments of a bacterial cell and the incorporation of this DNA molecule into the recepient chromosome in a heritable form.
In 1928 Frederick Griffith performed the experiments on Bacterial transformation with Streptococcus pneumonia the bacterium causing pneumonia. During the course of his experiment he found that a living organism (bacteria) could change in physical form.
- He observed two strains of this bacterium, one forming smooth colonies with capsule (S-type) and the other forming rough colonies without capsule (R-type).
- The S-type cells are virulent while R-type cells are non-virulent.
- When live S-type cells are injected into the mice, they suffered from pneumonia and died.
- When live R-type cells are injected into the mice, the disease did not appear and the mice survived.
- When heat killed S-type cells were injected, the disease did not appear.
- When heat killed S-type were mixed with live R-type cells and injected into the mice, the mice died of pneumonia and live S-type cells were isolated from the body of mice.
- He concluded that the R-strain had some how been transformed by the heat killed S- strain bacteria, which must be due to the transfer of genetic material the transforming principle.
![]()
Question 2.
Who revealed the biochemical nature of the transforming principle? How was it . done? Oswald Avery, Colin Mac Lead, and Madyn Me carty.
Answer:
Avery Mac Lead and Me carty revealed the biochemical nature of the transforming principle in a Griffith experiment.
- They purified biochemicals like proteins, DNA and RNA from the heat killed S-cells to see which one could transform live R cells into S cells.
- When their fraction were added to the culture of live R-cells, DNA was able to cause transformation of R-cells into S-cells.
- They also found that protein digesting enzymes and RNA digesting enzymes did not affect transformation indicating that transforming substance is not a protein or RNA.
- Digestion with DNase did inhibit transformation, this suggests that the DNA cause transformation.
Question 3.
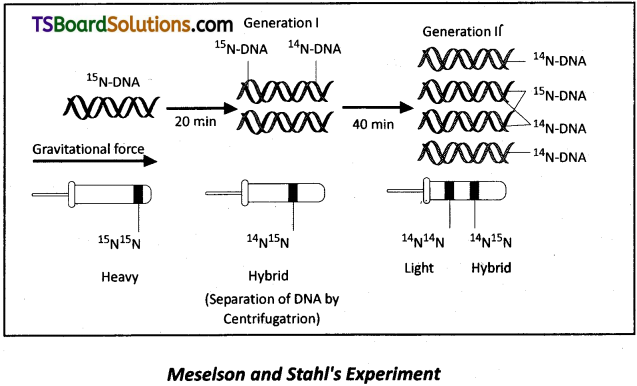
Discuss the significance of heavy isotope of nitrogen in Meselson and Stahl’s experiment.
Answer:
a) Matchew Meselson and Franklin Stahl performed an experiment using Escherischia coli to prove that DNA replication is semi conservative.
b) They grew E.coli in a medium containing 15NH4Cl (15N is the heavy isotope of nitrogen) as the only nitrogen source for many generations. This heavy DNA can be separated from the normal DNA by centrifugation in Cesium Chloride (CsCl) density gradient (Note that 15N is not a radioactive isotope and it can be separated from 14N based on densities only).
c) Then they transferred the cells into a medium with normal 14NH4Cl and took out samples at various time intervals and extracted DNA and centrifuged them to measure their densities.
d) Thus the DNA that was extracted from the culture one generation after transfer from 15N to 14N medium (that is after 20 minutes: E.coli divides in 20 minutes) had a hybrid or intermediate density.
e) The DNA extracted after two generations (i.e., after 40 minutes) consisted of equal amounts of light DNA and hyrbid DNA.
Question 4.
Define a cistron. Differentiate between monocistronic and polycistronic transcription unit with suitable examples. [May 2014]
Answer:
Cistron is defined as a segment of DNA coding for a polypeptide.
Monocistronic transcription :
Mostly in eukaryotes. The structural gene only one transcription unit can translate only one protein chain (or) one polypeptide chain. So it be said to be monocistronic transcription.
Polycistronic transcription:
Mostly in prokaryotes. The structural gene in a transcription unit can translate many polypeptides be said to be polycistronic transcription.
Question 5.
Recall the experiments done by Frederick Griffith in which DNA was speculated to be the genetic material. If RNA, instead of DNA, was the genetic material, would the heat killed strain of Pneumococcus have transformed the R-strain into virulent strain? Explain.
Answer:
- Stability as one of the properties of genetic material was clearly evident in Griffith’s transforming principle.
- Heat which killed bacteria, at least did not destroy some properties of genetic material.
- This can be explained by DNA. The two strands, being complementary, if separated by heating, come together when appropriate conditions are provided.
- RNA is has 2′ – OH group present at every nucleotide. It make RNA labile and easily degradable, RNA is reactive.
- RNA cannot be genetic material as RNA is degradable to heat, it cannot transform R strain into virulent strain.
![]()
Question 6.
There is only one possible sequence of amino acids when deduced from a given nucleotides. But a multiple nucleotide sequence can be deduced from a single amino acid sequence. Explain this phenomenon.
Answer:
AUG UUU UUC UUC UUU UUU UUC the only possible sequence of amino acid is
Met Phe Phe Phe Phe Phe Phe
Similarly from the sequence of following amino acids coded by an mRNA. Met-Phe-Phe-Phe-Phe-Phe-Phe the nucleotide sequence may be any one of UUU or UUC because these two codon code for Phenylalanine (Phe). As there are 64 codons for 20 amino acid and in which 3 codon do not code for any amino acids. Remaining codons must code for 20 amino acids. Therefore there will be more nucleotide for a single amino acid.
Question 7.
A single base mutation in a gene may not always result in loss or gain of function. Do you think the statement is correct? Defend your answer.
Answer:
No, the statement is not correct.
A classical example of point mutation is a change of single base pair in a gene for beta globin chain (in human haemoglobin) that results in the change of amino acid residue glutamate to valine. It results in a diseased condition called sickle cell anemia.
Thus a change in a single nucleotide in a codon alters the amino acid in a polypeptide chain.
For example :
Consider a statement that is made up of following words each having three letters like a genetic code.
RAM HAS RED CAP
If we insert a letter B in between HAS and RED and rearrange the statement it would read as follows.
RAM HAS BRE DCA P
The same can be repeated by deleting the one letter R then it will be the statement to make a triplet word.
RAM HAS EDC AP
Insertion or deletion of one or two bases changes the reading frame from the point of insertion or deletion. It can disrupt the protein structure and affect the functioning.
Question 8.
A low level of expression of lac operon occurs all the time. Can you explain the logic behind this phenomenon.
Answer:
- In lac operon (here lac refers to lactose), a polycistronic structural gene is regulated by a common promoter and regulatory genes. Such an arrangement is very common in bacteria and is referred to as operon.
- Lactose is the substrate for the enzyme beta-galactosidase and it regulates switching on and off of the operon. Hence it is termed as inducer.
- In the absence of a preferred carbon source such as glucose, if lactose is provided in the growth medium of the bacteria. It is transported into the cells through the action of permease.
- A very low level of expression of lac operon has to be present in the cell all the time, otherwise, lactose cannot enter the cells.
Question 9.
What background information did Watson and Crick have for developing a model of DNA? What was their contribution?
Answer:
Background information for developing a model of DNA was : DNA as an acidic substance present in the nucleus was first identified by Friedrich Meischer in 1869. He named it “Nuclein”. However, due to technical limitation in isolating such a long polymer intact, the elucidation of structure of DNA remained elusive for a long period of time.
It was in 1953 that James Watson and Francis Crick, based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, proposed a very simple and famous Double Helix model for the structure of DNA.
One of the hallmarks of their proposition was base pairing between the two strands of polynucleotide chains. However, this proposition was also based on the observation of Erwin Chargaff that for a double stranded DNA, the ratio between Adenine and Thymine and that between Guanine and Cytosine are constant and each equal ones.
Question 10.
What are the functions of
i) methylated guanosine cap,
ii) poly-A “tail” in a mature on RNA?
Answer:i) In capping and unusual nucleotide (methylated guanosine cap) is added to the 5′ end of hn RNA.
ii) In tailing, adenylate (Poly-A-tail) residues are added at 3’ end in the template independent manner. It is a fully processed hn RNA, now called mRNA.
Question 11.
How many types of RNA polymerases exist in cells? Write their names and functions.
Answer:
There are 3 types of RNA polymerases in the nucleus (in addition to the RNA polymerase found in the organelles).
They are
- RNA polymerase I transcribes rRNAs (28s, 18s & 5.8s)
- RNA polymerase II transcribes the precursor of mRNA, the heterogenous nuclear RNA (hn RNA).
- RNA polymerase III. It is responsible for transcription of tRNA < 5sr RNA and sn RNAs (Small nuclear RNAs).
![]()
Question 12.
Write briefly about DNA polymerase.
Answer:
DNA polymerase is the main enzyme in the replication of DNA.
It uses a DNA template to catalyse the polymerisation of deoxynucleotides only, in one direction i.e., 5′ → 3′ leading to one strand replication continuous and the other one as discontinuous.
The DNA polymerase can not initiate the process of replication on their own. A small stretch of RNA (called a primer) is required for initiation.
Question 13.
What are the contributions of George Gamow, H.G. Khorana, Marshall Nirenberg in deciphering the genetic code?
Answer:
George Gamow, a physicist argued that since there are only 4 bases and if they have to code for 20 amino acids, the code should constitute a combination of bases. He suggested that in order to code for all the 20 amino acids, the code should be made up of three nucleotides. This was a bold proposition, because a permutation and combination of 43 (4 × 4 × 4) would generate 64 codons, generating more codons than required.
Har Gobind Khorana developed a chemical method in synthesising RNA molecules with defined combinations of bases (homopolymers such as UUU and copolymers such as UUC, CCA).
Marshall Nirenberg’s made cell free system for protein synthesis. It finally helped the code to be deciphered.
Question 14.
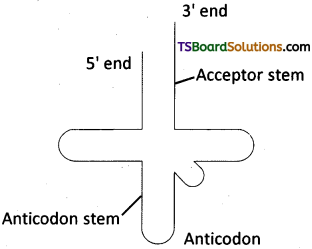
On the diagram of the secondary structure of tRNA shown below indicate the location of the following features :
a) Anticodon b) Acceptor stem c) Anticodon stem d) 5′ end e) 3′ end
Answer:
Question 15.
If there are 2.9 × 109 complete turns in a DNA molecule estimate the length of the molecule.
(1 angstrom = 10-8 cm).
Answer:
Distance between two consecutive base pair is 0.34 nm (0.34 × 10-9 m)
Complete turns in DNA = 2.9 × 109
Length of the DNA = Total no. of multiply with distance
between two consecutive bp = 2.9 × 109 × 0.34 × 10-9 = 0.986 m
Question 16.
Draw the schematic / diagrammatic presentation of the lac operon.
Answer:
Question 17.
What are the differences between DNA and RNA? [March ’20, ’14]
Answer:
| DNA | RNA |
| 1. It has two strands of nucleotides. | 1. It has only one strand of nucleotides. |
| 2. Most of the DNA is present in the nucleus and very little in chloroplast and mitochondria. | 2. Most of the RNA is present in cytoplasm and little in the nucleus. |
| 3. Deoxyribose sugar is present (C5H10O4). | 3. Ribose sugar is present (C5H10O5). |
| 4. Pyrimidines are Thymine and Cytosine. | 4. Pyrimidines are Uracil and Cytosine. |
| 5. DNA is made up of several nucleotides (more than 4 millions). | 5. RNA is made up of few nucleotides (75 – 2000 or more in mRNA). |
| 6. DNA undergoes self replication. | 6. Does not undergo self replication except in RNA viruses. |
| 7. DNA is the genetic material. | 7. RNA is non-genetic material (except in . RNA viruses). |
| 8. DNA does not participate directly in protein synthesis. | 8. RNA participates directly in protein synthesis. |
| 9. Metabolically DNA is of one type. | 9. Metabolically RNA is of three types. |
| 10. The base pairing is A = T and G ≡ C | 10. The base pairing is A = U and G ≡ C. |
![]()
Question 18.
Write the important features of Genetic code. [Mar. 18, 17]
Answer:
The important features of Genetic code are
- The codon is Triplet. 61 codons code for amino acids and 3 codons do not code for any amino acids, hence they function as stop codons.
- One codon codes for only one amino acid, hence it is unambiguous and specific.
- Some amino acids are coded by more than one codon, hence the code is degenerate.
- The codon is read in mRNA in a contiguous fashion. There are no punctuations.
- The code is nearly universal. For example, from bacteria to human, UUU would code for pheylalaline (Phe). Some exceptions to this rule have been found in mitochondrial codons, and in some protozoans.
- AUG has dual functions. It codes for Methionine (Met) and also acts as the initiotor codon.
Question 19.
Describe the sequential steps in the replication of a DNA molecule.
Answer:
- The process of replication involves a number of enzymes / catalysts of which DNA- dependent DNA-polymerase, is the major enzyme. It catalyses the polymerisation of the deoxy-ribonucleotides approximately at a rate of 2000 bs/second.
- The interwined DNA strand start separating from a particular point called origin of replication (which is single is prokaryotes and many in eukaryotes).
- This unwinding is catalysed by enzymes called helicases.
- Enzymes called topoisomerases break and reseal one of the strands of DNA, so that the unwind strands will not wind back.
- It is easy to add the base onto an already existing chain called primer.
- Primer is a short strech of RNA formed on the DNA template catalysed by enzyme primase.
- When a double stranded DNA is unwind upto a point, it shows a Y-shaped structure called replication fork.
- As new strands grow from the fork, it appears as if the point of divergence at the fork is moving.
- Enzyme DNA polymerase catalyses the joining of nucleotides (A, T, G, C) in the 5′ → 3′ direction.
- The enzymes forms one new strand in a continuous stretch (leading strand) in the 5′ → 3′ direction, on one of the template strands.
- On the other template strand, the enzyme forms short stretches of DNA (Okazaki fragments) also in the 5′ → 3′ direction.
- The Okazaki fragments are later joined by DNA – ligase to form the lagging strand.
- In prokaryotes, the DNA-polymerase does proof reading by removing any wrong base added, before proceeding to add new bases.
- The two strands are held together by hydrogen bonds between nucleotides.
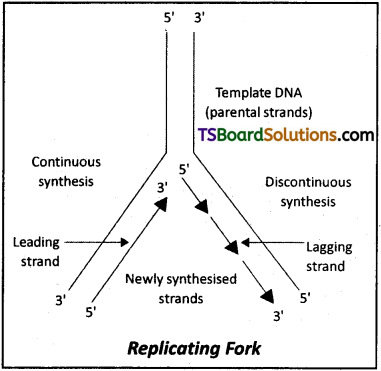
Question 20.
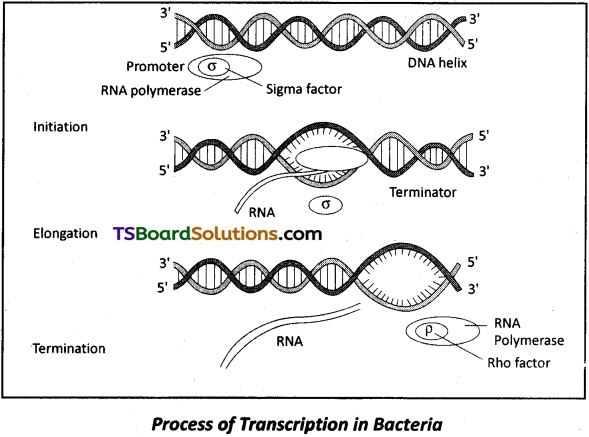
Give a diagrammatic presentation of the process of transcription in a bacterial cell.
Answer:
Question 21.
Write briefly on nudeosomes. [Mar. 2019, May ’17]
Answer:
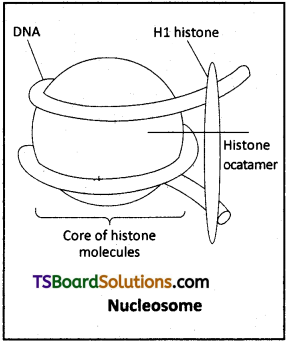
In DNA of eukaryotes, there are a set of positively charged basic proteins called histones.
- Histones are organised to form a unit of eight molecules called histone octamer.
- The negative charged DNA is wrapped around the positively charged histone octamer to form a structure called nudeosome.
- A typical nucleosome contains 200 bp of DNA helix.
- Nucleosome constitute the repeating unit of a structure in nucleus called chromatin.
- The nudeosomes in chromatin are seen as “beads-on-string” when viewed under electron microscope.
Long Answer Type Questions
Question 1.
Give an account of the Hershey and Chase experiment. What did it conclusively prove? If both DNA and proteins contained phosphorus and sulphur do you think the result would have been the same.
Answer:
Harshey and Chase Experiment:
- Their experiment is to prove that DNA is the genetic material and not the protein.
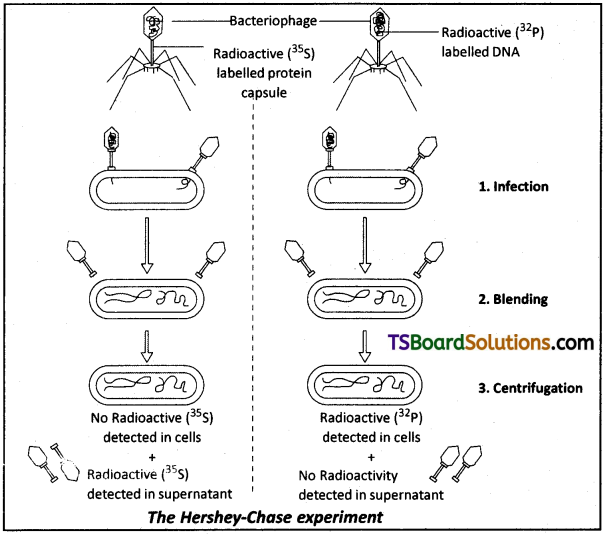
- They worked with bacteriophage T2 which attacks bacterium E.coli.
- The grew some viruses on the medium that contained radioactive phosphorus and some others on a medium that contained radioactive sulphur.
- Virus grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not.
- Similarly, virus, grown on radioactive sulphur contained radioactive protein but not radioactive DNA because DNA does not contain sulphur.
- Radioactive phages were allowed to attach to E.coli bacteria.
- The infection proceeded, the viral coats were removed from the bacteria by agitating them in a blender. The virus particles were separated from the bacteria by spinning them in a centrifuge.
- Bacteria which were infected with viruses that had radioactive DNA were radioactive, indicating that DNA was the material that passed from the virus to the bacteria.
- Bacteria that were infected with viruses that had radioactive proteins were not radioactive.
- This indicates that proteins did not enter the bacteria from the viruses.
- Thus it proves that DNA is the genetic material that is passed from virus to bacteria.
- If both DNA and proteins contained phosphorus and sulphur the result would not be same.
![]()
Question 2.
Give an account of post transcriptional modifications of a eukaryotic mRNA.
Answer:
- The primary transcripts contain both exons and introns and they are non-functional. Hence they are subjected to a process called splicing where the introns are removed and exons are joined in a defined order.
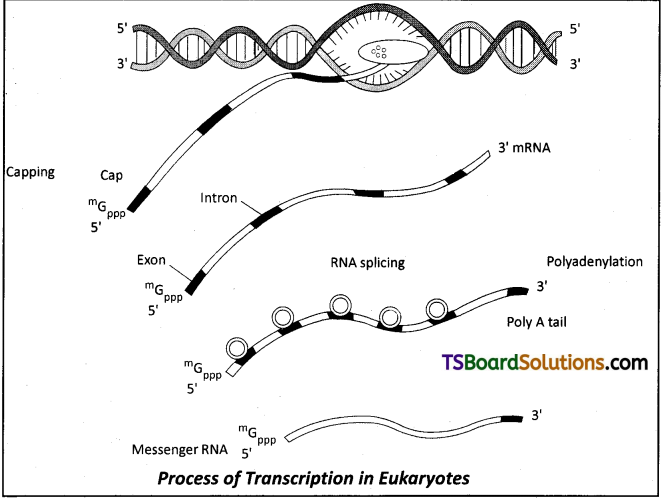
- hn RNA undergoes additional processing called capping and tailing, ft In capping an unusual nucleotide (methyl guanosine triphosphate) is added to the 5′ end of hn RNA.
- In tailing, adenylate residues (200-300) are added at 3′ end in a template independent manner.
- It is the fully processed hn RNA, now called mRNA, that is transported out of the nucleus for translation.
Question 3.
Discuss the process of translation in detail.
Answer:
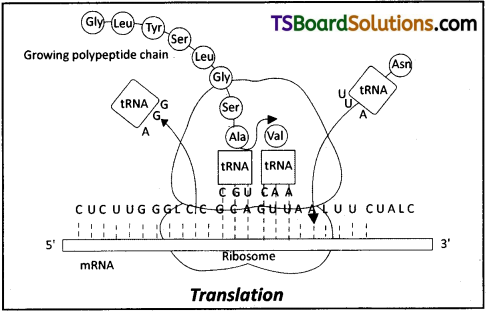
Translation is the process of polymerization of amino acids to form a polypeptide.
i) The amino acids are joined by a bond which is known as a peptide bond. This process requires energy.
ii) The different phases of translation are
a) Activation of amino acids
b) Initiation of polypeptide synthesis
c) Elongation of polypeptide chain
d) Termination of polypeptide chain
a) Activation of amino acids :
It occur in presence of ATP and linked to their cognate tRNA i.e., charging of tRNA or aminoacylation of tRNA. If two such charged tRNA, are brought close, the formation of peptide bond between them would occur energitically in presence of a catalyst.
b) Initiation of polypeptide synthesis occurs in ribosomes (cellular factory for protein synthesis):
- Ribosome consists of structural RNAs and about 80 different proteins.
- In its inactive state, it exists as two subunits – a large and a small subunit.
- When the small subunit encounters an mRNA, the process of translation of the mRNA to protein begins P-site and A-site.
- There are two sites in the large subunits – the P-site and A-site for subsequent aminoacids to bind to and thus close enough to each other for the formation of a peptide bond.
- The small subunit attaches to the large subunit in such a way that the initiation (AUG) comes to the P-site.
Elongation of polypeptide chain :
When a second tRNA charged with an appropriate amino acid binds to the A-site of the ribosome.
i) The peptide bond (CO – NH) forms between the carboxyl group of methionine and the amino group of the second amino acid. The reaction is catalysed by the enzyme peptidyl transferase.
ii) The complexes composed of an amino acid linked to tRNA, sequentially bind to the appropriate codon in mRNA by forming complementary base pairs with the tRNA anticodon.
iii) The ribosome moves from codon to codon along with the mRNA. Amino acids are added one by one, translated into polypeptide sequence dictated by DNA and represented by mRNA.
Termination of polypeptide synthesis occur when a released factor binds to the stop codon. As a result, the polypeptide synthesis or elongation stops releasing the complete polypeptide from the ribosome.
Question 4.
Write briefly about Griffith’s experiments on S. pneumoniae bacteria. What was his conclusion?
Answer:
Frederick Griffith (1928) performed the experiments on Bacterial transformation with Streptococcus pneumoniae, the bacterium causing pneumonia.
- He observed two strains of this bacterium, one forming smooth colonies with capsule (S-type) and the other forming rough colonies without capsule (R-type).
- The S-type cells are virulent while the R-type cells are non-virulent.
- When live S-type cells were injected into the mice, they suffered from pneumonia and died.
- When live R-type cells were injected into the mice, the disease did not appear and the mice survived.
- When heat killed S-type cells were injected, the disease did not appear.
- When heat killed S-type cells were mixed with live R-type cells and injected into the mice, the mice died of pneumonia and live S-type cells were isolated from the body of the mice.
- He concluded that the R-strain had somehow been transformed by the heat killed S-strain bacteria, which must be due to transfer of genetic material, the transforming principle.
Question 5.
Define an operon, giving an example. Explain what is an Inducible operon.
Answer:
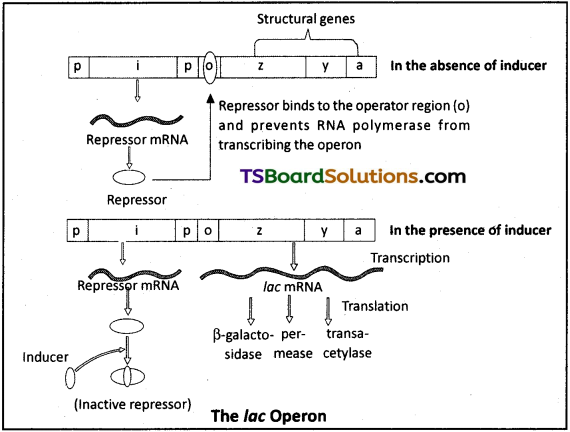
Operon is a group of closely packed structural genes and regulatory elements (DNA sequences) such as a regulator, a promoter and an operator, which function in a coordinated manner.
F. Jacob and J. Monod were first described a transcriptionally regulated system.
The lac operon (here lac refers to lactose) consists of one regulatory gene (i gene), the promoter (p) and operator (o) and the structural genes (z, y and a).
i) i – codes for repressor of the operon
z – codes for beta galactosidase (β-gal) y – codes for enzyme permease
a – codes for transacetylase enzyme ‘
ii) All the three gene products in lac operon are required for metabolism of lactose.
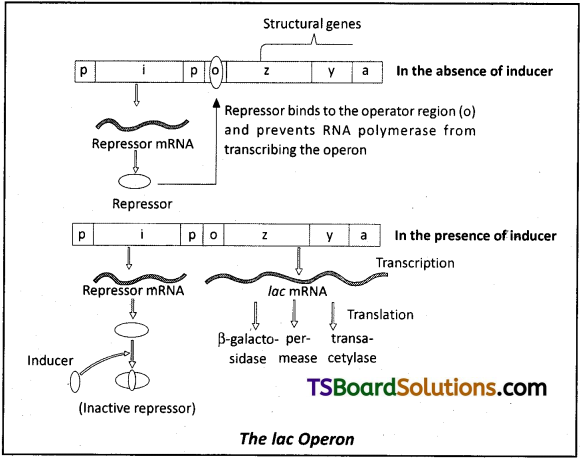
iii) Lactose is the substrate for the enzyme betagalactosidase and its regulates switching on and off of the operon. Hence it is termed as inducer.
iv) The lactose induces operon in the following way.
a) Repressor of the operon is synthesized from the i-gene.
b) Repressor protein binds to the operator region of the operon and prevents RNA polymerase from transcribing the operon.
c) In presence of an inducer, such as lactose or allolactose, the repressor is inactivated by the interaction with inducer. This allows RNA polymerase access to the promoter and transcription proceeds.
v) Regulation of lac operon by repressor is referred to as negative regulation.
![]()
Question 6.
Give the salient features of the Double helix structure of DNA.
Answer:
The salient features of a Double-Helix structure of DNA are as follows.
- It is made of two polynucleotide chains, where the backbone is constitued by sugar- phosphate and the bases project inside.
- The two chains have anti – parallel polarity. This means that if one chain has the polarity 5′ → 3′, the other has 3′ → 5′.
- The bases in two strands are paired through hydrogen bonds (H-bonds) forming base pairs (bp). Adenine forms two hydrogen bonds with Thymine from the opposite strand and vice-versa. Similarly Guanine is bonded with Cytosine with three hydrogen bonds. As a result, a Purine always comes opposite to a Pyramidine. This generates approximately uniform distance (20 A) between the two strands of the helix.
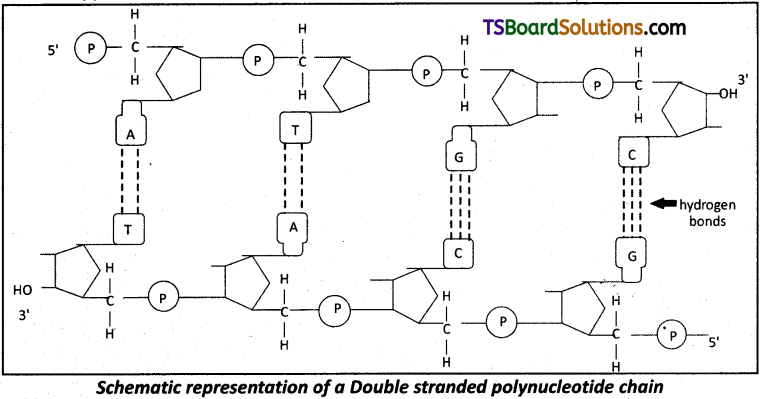
- The two chains are coiled in a right handed fashion. The pitch of the helix is 3.4 nm a nanometer is one billionth of a metre, that is 10-9 m) and there are roughly 10 bp in each turn. Consequently, the distance between two successive base pairs (bp) in a helix is approximately equal to 0.34 nm.
- The plane of one base pair stacks over the other in a double helix. This, in addition to H-bonds, confers stability of the helical structure.
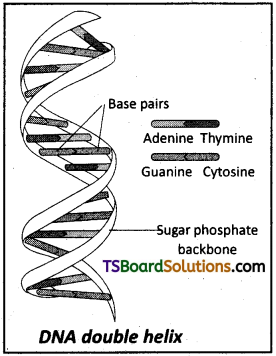
Question 7.
Replication was allowed to take place in the presence of radioactive Deoxy-nucleotide precursors in E.coli that was a mutant for DNA ligase. Explain how the newly synthesised radioactive DNA will be when purified.
Answer:
- In the long DNA molecule, the replication occurs within a small opening of DNA Helix, called Replication fork.
- On one strand, the template with polarity (3′ – 5′) the replication is continuous.
- On the other strand, the template with polarity (5′ – 3′), it would be discontinuous.
- The discontinuously synthesized fragments are joined by the DNA ligase.
- When Replication was allowed to take place in the presence of radioactive Deoxy – nucleotide precusors in E.Coli that was a mutant for DNA ligase, their Okazaki fragments will not be joined.
Intext Question Answers
Question 1.
Group the following as nitrogenous bases and nucleosides: Adenine, Cytidine, Thymine, Guanosine, Uracil and Cytosine.
Answer:
Nitrogenous bases present in the list are adenine, thymine, uracil and cytocine. Nucleosides present in the list are cytidine and guanosine.
Question 2.
If a double stranded DNA has 20 per cent of cytosine, calculate the percent of adenine in the DNA.
Answer:
According to Chargaff’s rule, the DNA molecule should have an equal ratio of pyrimidine (cytocine and thymine) and purine (adenine and guanine). It means that the number of adenine molecules is equal to thymine molecules and the number of guanine molecules is equal to cytosine molecules.
% A = % T and % G = % C
If ds DNA has 20% of cytosine then according to the law, it would have 20% of guanine. Thus percentage of G + C content = 40%. The remaining 60% represents both A + T molecules. Since adenine and guanine are always present in equal number the percentage of adenine molecule is 30%.
![]()
Question 3.
If the sequence of one strand of ONA is written as follows :
Write down the sequence of complementary strand in 3′ → 5′ direction.
5′ – ATGCATGCATGCATGCATGCATGCATGC – 3′
Answer:
The DNA strand are complementary to each other with respect to base sequence. Hence if the sequence of one strand of DNA is
5′ ATGCATGCATGCATGCATGCATGCATGC -3′
Then the sequence of complementary strand in
3′ TACGTACGTACGTACGTACGTACGTACG – 5′ direction.
Therefore the sequence of nucleotides in DNA polypeptide direction in 5′ – GCATGCATGCATGCATGCATGCATGCAT 3′.
Question 4.
If the sequence of the coding strand in a transcription unit is written as follows : 5′ – ATGCATGCATGCATGCATGCATGCATGC – 3′. Write down the sequence of mRNA.
Answer:
If the coding strand in a transcription unit is 5′ ATGCATGCATGCATGCATGCATGC ATGC 3′ Then the template strand in 3′ to 5′ direction would be 3′ TACGTACGTACGTACGTACGTACGTACG 5′
It is known that the sequence of mRNA is same on the coding strand of DNA. However, in RNA, thymine is replaced by uracil. Hence the sequence of mRNA will be 5′ AUGCAUGCAUGC AUGC AUGC AUGC – 3′
Question 5.
Which property of DNA double helix led Watson and Crick to hypothesise semiconservative mode of DNA replication? Explain.
Answer:
Watson and Crick observed that the two strands of DNA are anti parallel and complementary to each other with respect to their base sequences. This type of arrangement in DNA molecule led to the hypothesis that DNA replication is semiconservative. It means that the double stranded DNA molecule separates and then each of the separated strand acts as a template for the synthesise of a new complementary strand. As a result, each DNA molecule would have one parental strand and a newly synthesized daughter strand. Since only one parental strand is conserved in each daughter molecule. It is semi-conservative mode of replication.
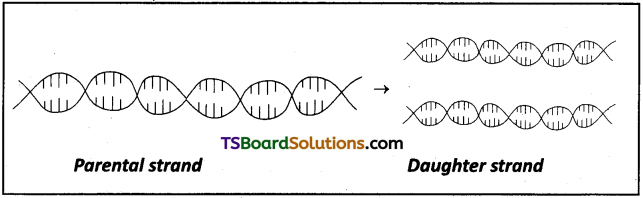
Question 6.
Depending upon the chemical nature of the template (DNA or RNA) and the nature of nucleic acids synthesized from it (DNA or RNA), list the types of nucleic acid polymerases.
Answer:
There are two different types of polymerases.
- DNA – dependent DNA polymerases.
- DNA dependent RNA polymerases.
The DNA dependent DNA polymerases use a DNA template for synthesizing a new strand of DNA. Where as DNA-dependent RNA polymerases use a DNA template strand for synthesizing DNA.
Question 7.
How did Hershey and Chase differentiate between DNA and protein in their experiment while proving that DNA is the genetic material?
Answer:
Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. Similarly viruses grown on radioactive sulphur contained radioactive protein but not radioactive DNA because DNA does not contain sulphur.
![]()
Question 8.
Differentiate between the followings :
a) mRNA and tRNA
b) Template strand and Coding strand.
Answer:
a) mRNA is a single stranded and unfolded polynucleotide molecule. It carries a genetic information required for the synthesis of a specific protein from DNA to ribosomes in the form of triplet codons.
tRNA is the smallest RNA also called soluble RNA (sRNA) or Adaptor RNA or Translator RNA. It is like clover leaf. It is single stranded which is folded forming three distinct loops and a 4th indistinct loop which is considered as accessory arm along with a tail.
b) DNA at promoter sites gives two single strands, one of which is called template strand which is transcribed where as the other strand is called coding strand which does not code for anything and is not transcribed.
Question 9.
List two essential roles of ribosome during translation.
Answer:
Ribosomes are responsible for protein synthesis. The ribosomes consists of structural RNAs and about 80 different proteins. In its inactive state, it exists as two subunits : a large subunit and a small subunit. When the small subunit encounter an mRNA, the process of translation of the mRNA to proteins begins. There afe two sites in the large subunit for subsequent amino acids to bind to and thus be close enough to each other for the formation of a peptide bond. The ribosome also acts as a catalyst for the formation of a peptide bond.
Question 10.
In the medium where E.coli was growing, lactose was added, which induced the lac operon. Then, why does lac operon shut down some time after addition of lactose in the medium?
Answer:
- Lactose is the inducer for lac operon.
- The active form of lactose binds to the repressor protein and brings about a conformational change in the repressor.
- As a result the repressor is inactive i.e., it cannot bind to the operator.
- This provides access of the RNA polymerase to structural genes and transcription and production of enzymes continue and metabolism of lactose takes place.
- In its absence, the repressor is active and binds to the operator. There by switching off the process.
Question 11.
Explain (in one or two lines) the function of the followings :
a) Promoter b) t RNA c) Exons t
Answer:
a) Promoter :
It is a region of DNA where RNA polymerase binds and initiates transcription.
b) t RNA :
Transfer RNA is an adaptor molecule, that is used by all living organisms to bridge the genetic code in messenger RNA with the twenty amino acids in proteins. tRNA carry amino acids to ribosomes, where they are linked into proteins.
c) Exons :
In eukaryotes, the monocistronic structural genes have interrupted coding sequences – the genes in eukaryotes are split. The coding sequences or expressed sequences are defined as Exons. Exons are interrupted by introns.
Question 12.
Briefly describe the following:
a) Transcription
b) Translation
Answer:
a) Transcription :
Transfer of the genetic information from the DNA blue print to the mRNA is called transcription.
b) Translation :
Arrangement of aminoacids in a linear, specific sequence and formation of polypeptide chain according to the specific sequence of the information written on the m RNA is called translation.
Question 13.
How the polymerization of nucleotides can be prevented in a DNA molecule.
Answer:
DNA polymerases on their own cannot initiate the process of replication. By removing primer DNA template and absence of DNA polymerase.
Question 14.
In an experiment, DNA is treated with a compound which tends to place itself amongst the stacks of nitrogenous base pairs. As a result of this, the distance between two consecutive base pairs increases from 0.34 nm to 0.44 nm calculate the length of DNA double helix (which has 2 × 109 bp) in the presence of saturating amount of this compound.
Answer:
Distance between 2 consecutive base pair in 0.44 nm (0.44 × 10-9 m)
The length of DNA double helix = 6.6 × 2 × 109 bp × 0.44 × 10-9 m / bp = 5.8 metres.
Question 15.
Recall the experiments done by Frederick Griffith. Where DNA was speculated to be the genetic material. If RNA, instead of DNA was the genetic material, would the heat killed strain of Pneumococcus have transformed the R-strain into virulent strain? Explain.
Answer:
Heat, which killed the bacteria, at least did not destroy some of the properties of the genetic material.
Moreover 2′ – OH group present at every nucleotide in RNA is a reactive group and makes RNA liable and easily degradable.
RNA is known to be catalyst hence reactive.
So, If RNA instead of DNA was the genetic material, the heat killed strain of pseudococcus could not transform the R-strain into virulent strain.
Question 16.
You are repeating the Hershey-Chase experiment and are provided with two isotopes: 32P and 15N (in place of 35S in the original experiment). How do you expect your results to be different?
Answer:
Harshey and Chase worked to discover whether it was protein or DNA from the virus that entered the bacteria.
If we repeat the same experiment by using two isotopes 32P and 15N (in place of 35S in the original experiment) we cannot distinguish the difference between the protein or DNA.
32P and 15N both are present in DNA in the Bacteria which was infected with virus. But we can not distinguish that protein did not enter the bacteria.
Question 17.
Do you think that the alternate splicing of exons may enable a structural gene to code for serveral isoproteins from one and the same gene? If yes, how? If not, why so?
Answer:
Yes. A gene is split into several sections separated by non-coding segments of DNA. Such genes are called discontinuous genes or split genes.
In split genes there are two sections – introns and exons.
Introns are non-coding sections of DNA segment. Before translation, the introns have to be removed and the exons reattached to produce a final copy of mRNA with continuous codons. This is called gene splicing.
According to the one gene one protein hypothesis of Beadle’arnd Tatum, each gene will separately code for one protein and that genes do not overlap.
So alternative splicing of exons may enable a structural gene to code for several isoproteins from one and the same gene.
![]()
Question 18.
Can you recall what centrifugal force is, and think why a molecule with higher mass / density would sediment faster?
Answer:
Centrifugal force is the force that acts away from the centre of the circle. Higher mass / density will be thrown towards outside because heavier particles experience more centrifugal force. So they would sediment faster.
Question 19.
Do Retroviruses follow central Dogma? Give one example.
Answer:
Francis Crick proposed the central dogma in molecular biology, which states that genetic information flows from

Retroviruses do not follow central dogma.
Retroviruses contain RNA as genetic material flow information in the reverse direction, that is from RNA to DNA.
Example: HIV